A structural and mechanistic view on enzymes evolution
Research overview
Our contributions to the study of antibiotic resistance evolution!
Our group is dedicated to understanding the evolution of β-lactamases and biofilm-associated enzymes. Our long-term goal is to uncover the molecular mechanisms by which evolution modulates enzyme structure and activity and to leverage this knowledge to develop new compounds that target these enzymes effectively.
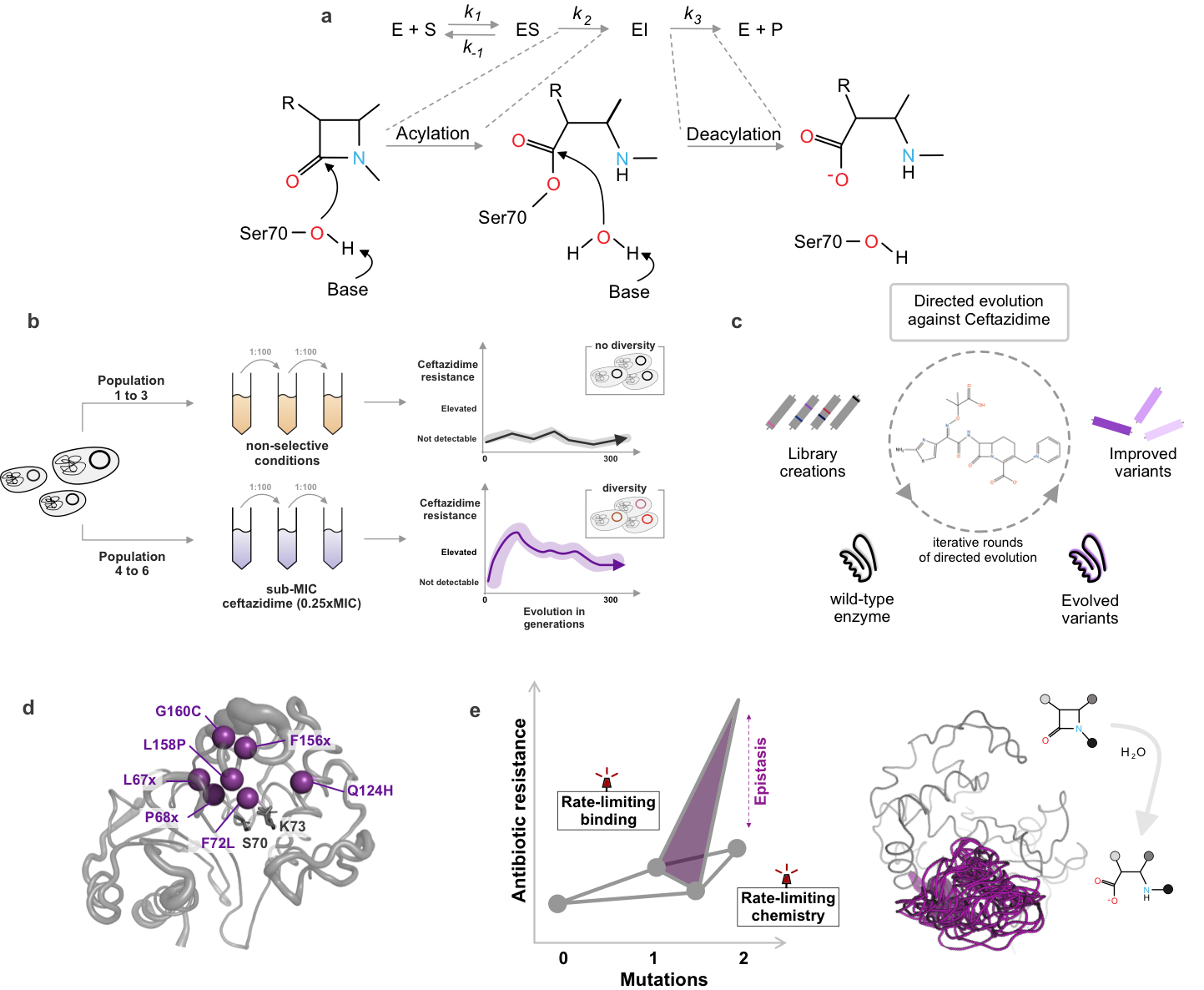
Overview of our work on the evolution of β-lactamases
(a) illustrates the general reaction mechanism of serine β-lactamases, enzymes that hydrolyze β-lactam antibiotics and confer resistance. To explore their evolutionary potential, we use a combination of long-term experimental evolution (b) and directed evolution approaches (c), enabling us to investigate how these enzymes adapt under different selective pressures. To uncover the molecular mechanisms driving these adaptations, our group integrates structural biology (d) and biochemistry (e). This interdisciplinary strategy allows us to link changes in enzyme sequence and structure to their functional consequences, shedding light on how resistance evolves and how it might be counteracted.
Figure: adapted from Frøhlich et al 2021 and Frøhlich et al 2024
Our Work on Inhibitor Design and Testing
Our research aims to discover and develop inhibitors targeting β-lactamases. Using a fragment-based drug discovery approach, we have screened over 500 small molecular fragments and identified several that inhibit key enzymes, including the metallo-β-lactamases VIM-2 and NDM-1, as well as the serine β-lactamase OXA-48. Guided by high-resolution crystal structures of our target enzymes, we apply structure-based design and molecular modelling to refine these fragment hits into larger molecules with improved inhibitory properties. The resulting compounds are evaluated using biochemical and cellular assays to assess potency and specificity, ranging from enzyme kinetics, surface plasmon resonance (SPR), and IC₅₀ determination to whole-cell assays measuring bacterial susceptibility.
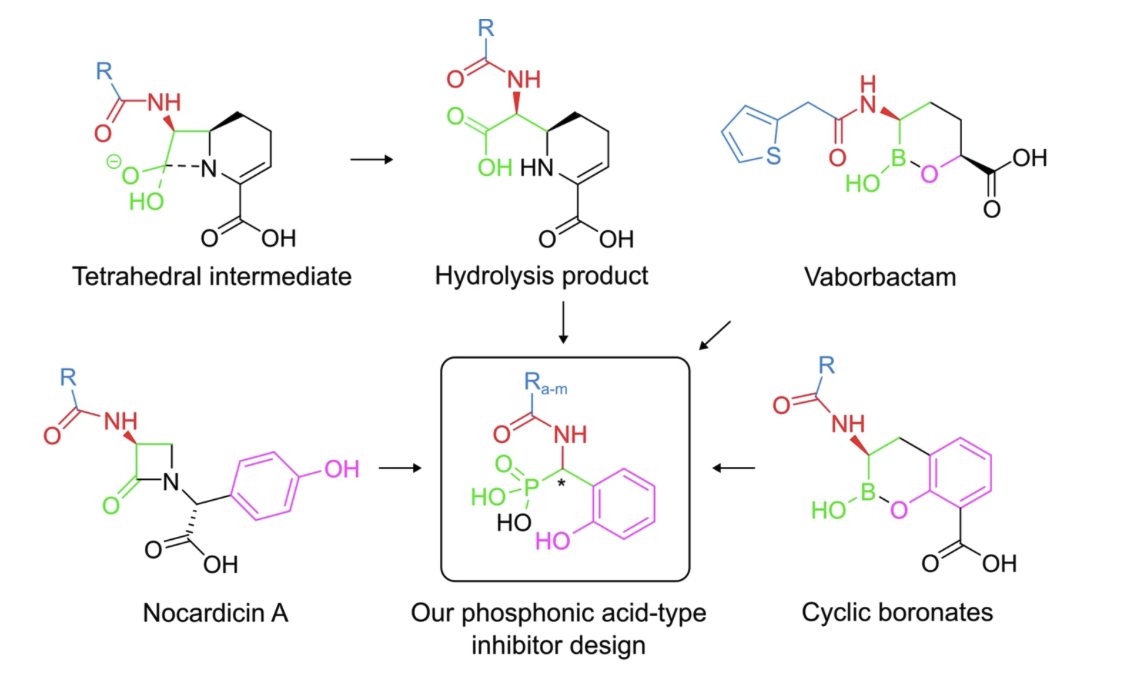
Showcase: Phosphonic Acid–Based MBL Inhibitors
Our PhD student Daniel Salamonsen, in collaboration with the Erdélyi group, developed and characterized phosphonic acid–based inhibitors targeting metallo-β-lactamase (MBL) enzymes. These dynamically chiral phosphonic acids are straightforward to synthesize, penetrate bacterial membranes, inhibit clinically relevant MBLs (NDM-1, VIM-2, and GIM-1), and show no toxicity toward human cells. By mimicking the transition state of β-lactam hydrolysis and coordinating the Zn ions in the enzyme active site, both stereoisomers can bind the enzyme, providing exceptional adaptability and the potential to slow resistance evolution.
Figure: adapted from Gulyás et al 2025
Check out our latest preprints!
Biofilm evolution in Vibrio cholerae
Øyvind Lorentzen discovered how biofilm evolution in Vibrio cholerae reshapes the signaling enzyme MbaA, disrupting its normal regulation and boosting levels of the messenger molecule c-di-GMP. These changes promote a shift from free-living to biofilm-associated lifestyles, revealing how evolution can reprogram cellular signaling.
Preprint on BioRxiv: Lorenzten et al 2025
Evolution of functional sub-states in the β-lactamase OXA-48
Daniel Salamonsen studied the mechanisms of how OXA-48 evolves to confer increased ceftazidime resistance. We show that evolution step-wise selected for a pre-exisitng sub-state. The interconversion rates between functional sub-states limited overall catalytic output. Evolution accelerates these processes, relieving kinetic bottlenecks in the conformational landscape and directly increasing ceftazidime hydrolysis.
Preprint on BioRxiv: Salamonsen et al 2025